Fluorine »
PDB 3syn-3u2o »
3ttz »
Fluorine in PDB 3ttz: Crystal Structure of A Topoisomerase Atpase Inhibitor
Enzymatic activity of Crystal Structure of A Topoisomerase Atpase Inhibitor
All present enzymatic activity of Crystal Structure of A Topoisomerase Atpase Inhibitor:
5.99.1.3;
5.99.1.3;
Protein crystallography data
The structure of Crystal Structure of A Topoisomerase Atpase Inhibitor, PDB code: 3ttz
was solved by
P.A.Boriack-Sjodin,
J.Read,
A.E.Eakin,
B.A.Sherer,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 51.74 / 1.63 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 143.178, 55.629, 51.216, 90.00, 100.37, 90.00 |
| R / Rfree (%) | 17.3 / 19.4 |
Other elements in 3ttz:
The structure of Crystal Structure of A Topoisomerase Atpase Inhibitor also contains other interesting chemical elements:
| Magnesium | (Mg) | 5 atoms |
| Chlorine | (Cl) | 4 atoms |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Crystal Structure of A Topoisomerase Atpase Inhibitor
(pdb code 3ttz). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 2 binding sites of Fluorine where determined in the Crystal Structure of A Topoisomerase Atpase Inhibitor, PDB code: 3ttz:
Jump to Fluorine binding site number: 1; 2;
In total 2 binding sites of Fluorine where determined in the Crystal Structure of A Topoisomerase Atpase Inhibitor, PDB code: 3ttz:
Jump to Fluorine binding site number: 1; 2;
Fluorine binding site 1 out of 2 in 3ttz
Go back to
Fluorine binding site 1 out
of 2 in the Crystal Structure of A Topoisomerase Atpase Inhibitor
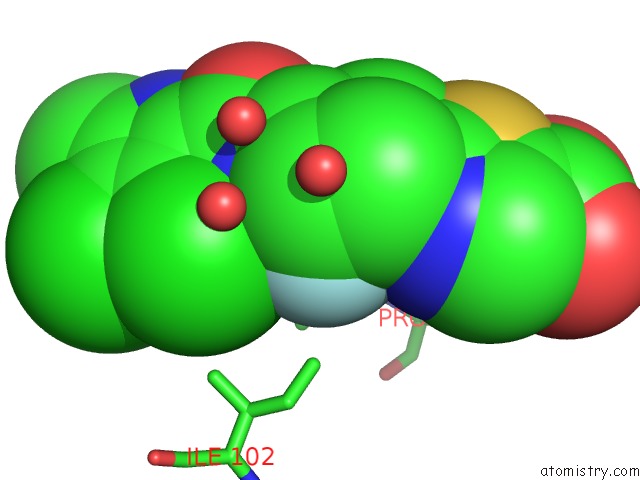
Mono view
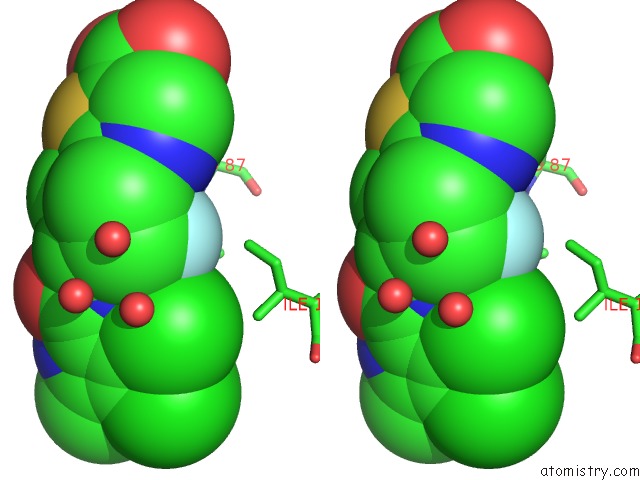
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Crystal Structure of A Topoisomerase Atpase Inhibitor within 5.0Å range:
|
Fluorine binding site 2 out of 2 in 3ttz
Go back to
Fluorine binding site 2 out
of 2 in the Crystal Structure of A Topoisomerase Atpase Inhibitor
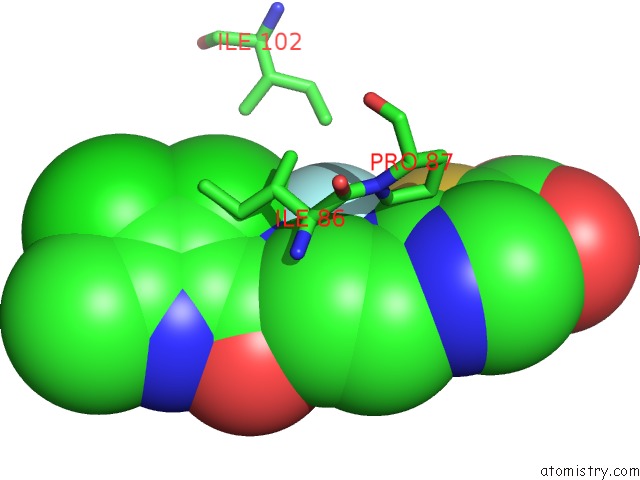
Mono view
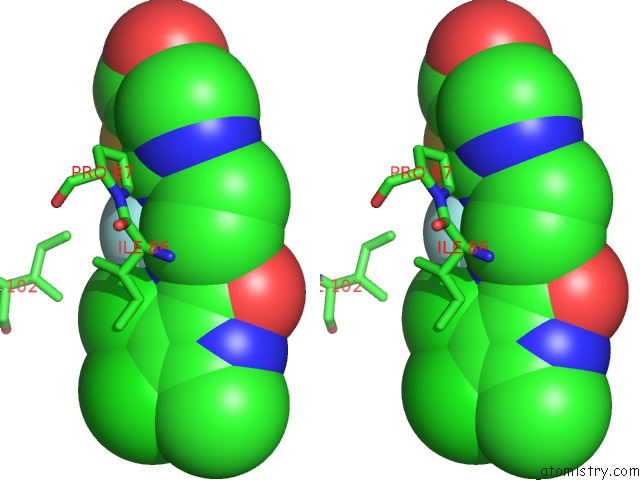
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Crystal Structure of A Topoisomerase Atpase Inhibitor within 5.0Å range:
|
Reference:
B.A.Sherer,
K.Hull,
O.Green,
G.Basarab,
S.Hauck,
P.Hill,
J.T.Loch,
G.Mullen,
S.Bist,
J.Bryant,
A.Boriack-Sjodin,
J.Read,
N.Degrace,
M.Uria-Nickelsen,
R.N.Illingworth,
A.E.Eakin.
Pyrrolamide Dna Gyrase Inhibitors: Optimization of Antibacterial Activity and Efficacy. Bioorg.Med.Chem.Lett. V. 21 7416 2011.
ISSN: ISSN 0960-894X
PubMed: 22041057
DOI: 10.1016/J.BMCL.2011.10.010
Page generated: Mon Jul 14 19:30:20 2025
ISSN: ISSN 0960-894X
PubMed: 22041057
DOI: 10.1016/J.BMCL.2011.10.010
Last articles
F in 4MM8F in 4MIH
F in 4MM4
F in 4MK0
F in 4MIG
F in 4MH0
F in 4MDD
F in 4MDB
F in 4MC9
F in 4MDA