Fluorine »
PDB 6p3v-6pf9 »
6p6g »
Fluorine in PDB 6p6g: Co-Crystal Structure of Human SMYD3 with Isoxazole Amides Inhibitors
Enzymatic activity of Co-Crystal Structure of Human SMYD3 with Isoxazole Amides Inhibitors
All present enzymatic activity of Co-Crystal Structure of Human SMYD3 with Isoxazole Amides Inhibitors:
2.1.1.43;
2.1.1.43;
Protein crystallography data
The structure of Co-Crystal Structure of Human SMYD3 with Isoxazole Amides Inhibitors, PDB code: 6p6g
was solved by
P.A.Elkins,
L.Wang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 56.61 / 1.59 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 61.433, 66.636, 107.340, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.2 / 20 |
Other elements in 6p6g:
The structure of Co-Crystal Structure of Human SMYD3 with Isoxazole Amides Inhibitors also contains other interesting chemical elements:
| Magnesium | (Mg) | 2 atoms |
| Zinc | (Zn) | 3 atoms |
Fluorine Binding Sites:
The binding sites of Fluorine atom in the Co-Crystal Structure of Human SMYD3 with Isoxazole Amides Inhibitors
(pdb code 6p6g). This binding sites where shown within
5.0 Angstroms radius around Fluorine atom.
In total 3 binding sites of Fluorine where determined in the Co-Crystal Structure of Human SMYD3 with Isoxazole Amides Inhibitors, PDB code: 6p6g:
Jump to Fluorine binding site number: 1; 2; 3;
In total 3 binding sites of Fluorine where determined in the Co-Crystal Structure of Human SMYD3 with Isoxazole Amides Inhibitors, PDB code: 6p6g:
Jump to Fluorine binding site number: 1; 2; 3;
Fluorine binding site 1 out of 3 in 6p6g
Go back to
Fluorine binding site 1 out
of 3 in the Co-Crystal Structure of Human SMYD3 with Isoxazole Amides Inhibitors

Mono view
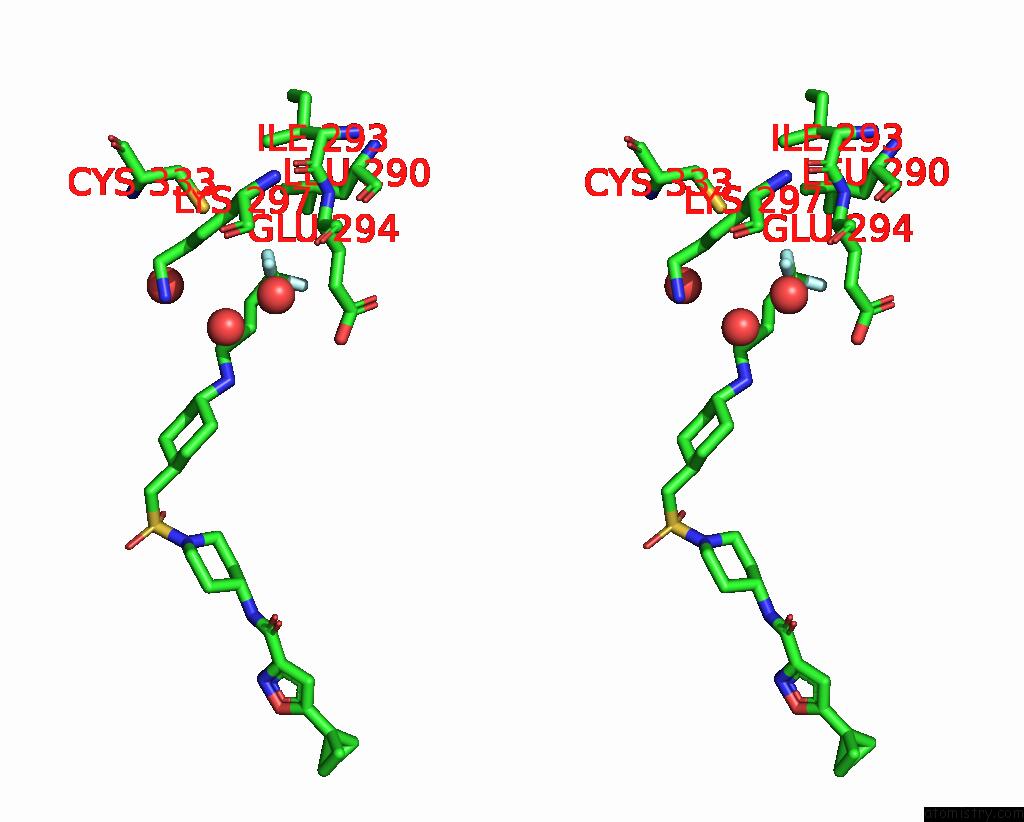
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 1 of Co-Crystal Structure of Human SMYD3 with Isoxazole Amides Inhibitors within 5.0Å range:
|
Fluorine binding site 2 out of 3 in 6p6g
Go back to
Fluorine binding site 2 out
of 3 in the Co-Crystal Structure of Human SMYD3 with Isoxazole Amides Inhibitors
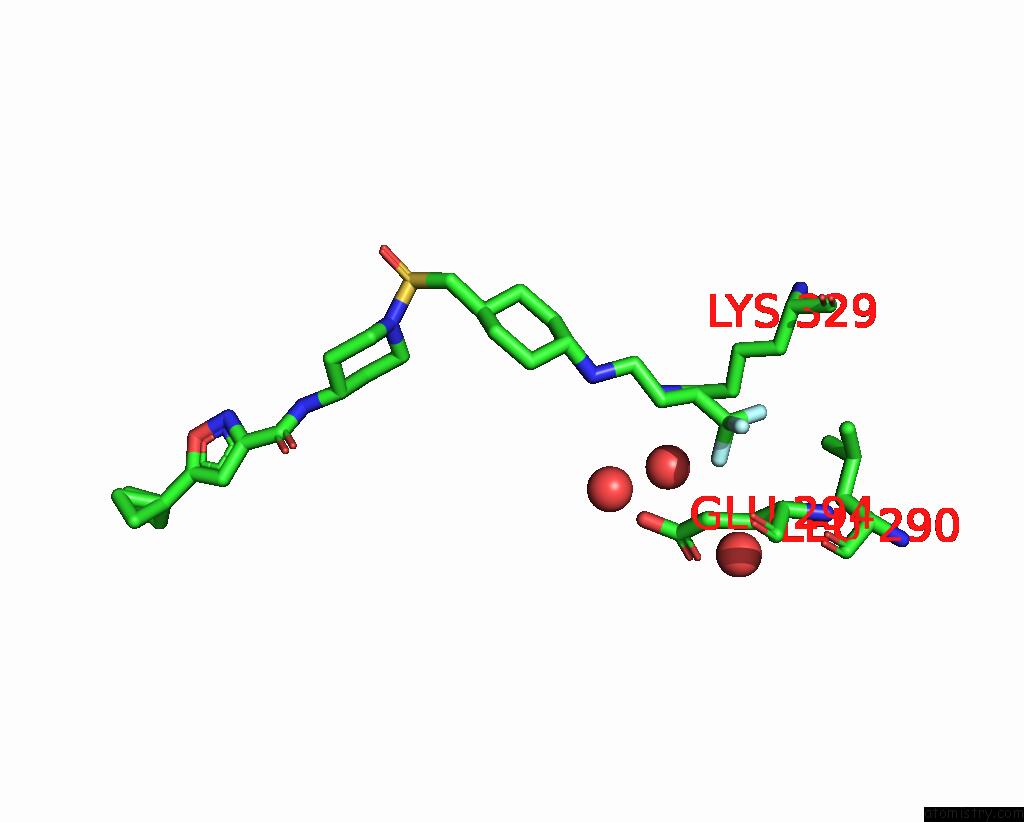
Mono view
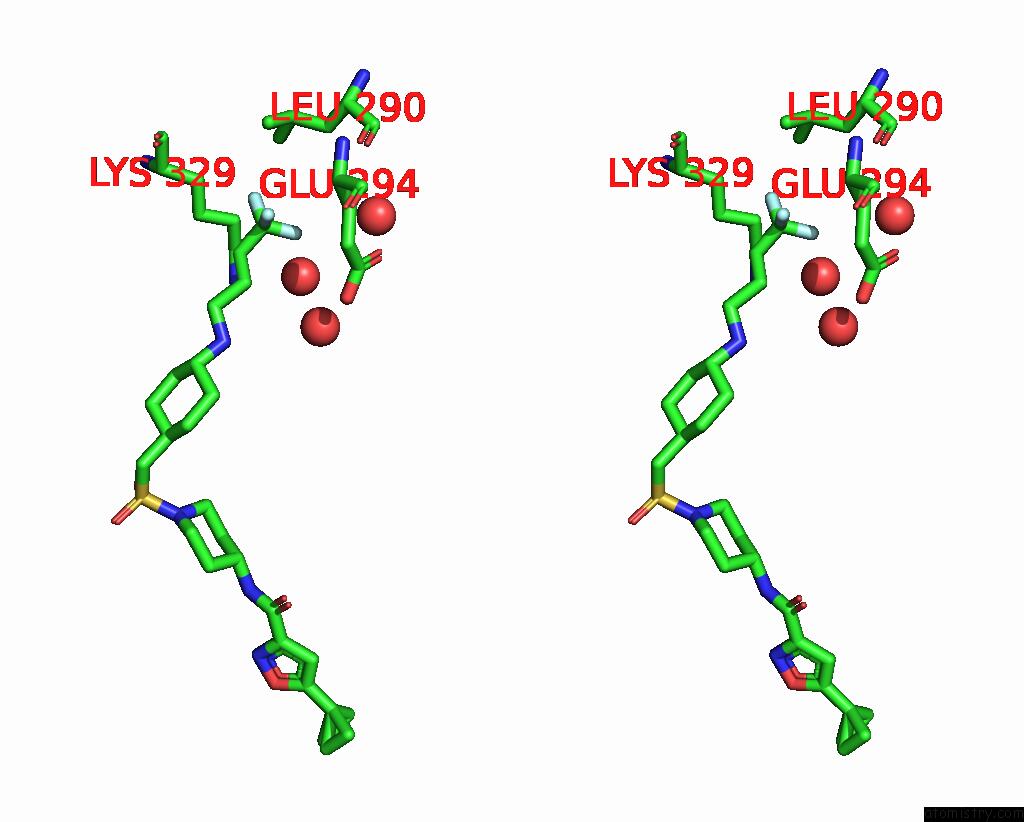
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 2 of Co-Crystal Structure of Human SMYD3 with Isoxazole Amides Inhibitors within 5.0Å range:
|
Fluorine binding site 3 out of 3 in 6p6g
Go back to
Fluorine binding site 3 out
of 3 in the Co-Crystal Structure of Human SMYD3 with Isoxazole Amides Inhibitors
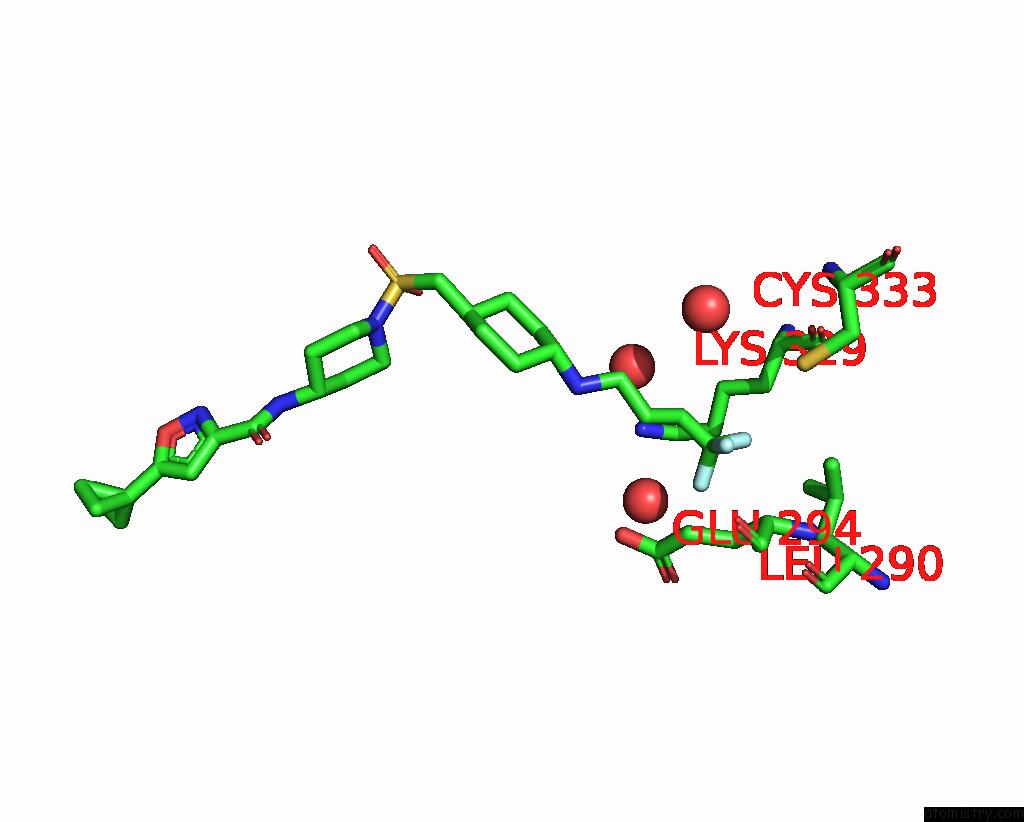
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Fluorine with other atoms in the F binding
site number 3 of Co-Crystal Structure of Human SMYD3 with Isoxazole Amides Inhibitors within 5.0Å range:
|
Reference:
D.-S.Su,
J.Qu,
M.Schultz,
C.W.Blackledge,
H.Yu,
J.Zeng,
J.Burgess,
A.Reif,
M.Stern,
R.Nagarajan,
M.Baker Pappalardi,
K.Wong,
A.P.Graves,
W.Bonnette,
L.Wang,
P.A.Elkins,
B.Knapp-Reed,
J.Carson,
C.Mchugh,
H.Mohammad,
R.Kruger,
J.Luengo,
D.A.Heerding,
C.L.Creasy.
Discovery of Isoxazone Amides As Potent and Selective SMYD3 Inhibitors Acs Med.Chem.Lett. 2019.
ISSN: ISSN 1948-5875
Page generated: Tue Jul 15 14:28:40 2025
ISSN: ISSN 1948-5875
Last articles
F in 7KHHF in 7KHG
F in 7KH6
F in 7KBG
F in 7KBN
F in 7KBC
F in 7KAF
F in 7KA1
F in 7K77
F in 7K8G